Signal Processor¶
After running a simulation, you may need to apply some post-simulation signal processing. For example, you may need to scale the simulated spectrum to match experimental intensities, or you may want to convolve the spectrum with a Lorentzian, Gaussian, or other line-broadening
function. For this reason, mrsimulator offers some frequently used NMR signal processing tools within the mrsimulator.signal_processor module.
See also
Signal Processing Gallery for notebooks using common processing functions.
CSDM object¶
The simulated spectrum is held in a CSDM [1] object, which supports multi-dimensional scientific datasets (NMR, EPR, FTIR, GC, etc.). For more information, see the csdmpy documentation.
SignalProcessor class¶
Signal processing is a series of operations sequentially applied to the dataset.
In mrsimulator, the SignalProcessor object is
used to apply operations. Here we create a new SignalProcessor object
# Import the signal_processor module
from mrsimulator import signal_processor as sp
# Create a new SignalProcessor object
processor = sp.SignalProcessor()
Each signal processor object holds a list of operations under the operations attribute. Below we add operations to apply Gaussian line broadening and a scale factor.
processor.operations = [
sp.IFFT(),
sp.apodization.Gaussian(FWHM="50 Hz"),
sp.FFT(),
sp.Scale(factor=120),
]
First, an inverse Fourier transform is applied to the dataset, converting
it to the time domain. Then, a Gaussian apodization, parameterized using a
full-width-at-half-maximum (FWHM) of 50 Hz, is applied. Note, the
dimensionality of the FWHM attribute has the inverse dimensionality
of the dataset domain. Finally, a forward Fourier transform is applied to
the apodized dataset, and all points are scaled up by 120 times.
Note
Convolutions in mrsimulator are performed using the Convolution Theorem. A spectrum is Fourier transformed, and apodizations are performed in the time domain before being transformed back into the frequency domain.
Let’s create a CSDM object and then apply the operations to visualize the results.
import csdmpy as cp
import numpy as np
# Create a CSDM object with delta function at 200 Hz
test_data = np.zeros(500)
test_data[200] = 1
csdm_object = cp.CSDM(
dependent_variables=[cp.as_dependent_variable(test_data)],
dimensions=[cp.LinearDimension(count=500, increment="1 Hz")],
)
To apply the previously defined signal processing operations to the above CSDM object, use
the apply_operations() method of the
SignalProcessor instance as follows
processed_dataset = processor.apply_operations(dataset = csdm_object)
The variable processed_dataset is another CSDM object holding the dataset
after the list of operations has been applied to csdm_object. Below is a
plot comparing the unprocessed and processed dataset
import matplotlib.pyplot as plt
_, ax = plt.subplots(1, 2, figsize = (8, 3), subplot_kw = {"projection":"csdm"})
ax[0].plot(csdm_object, color="black", linewidth=1)
ax[0].set_title("Unprocessed")
ax[1].plot(processed_dataset.real, color="black", linewidth=1)
ax[1].set_title("Processed")
plt.tight_layout()
plt.show()
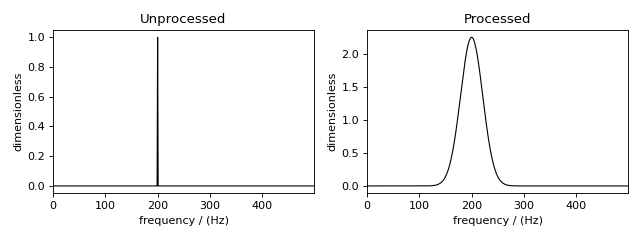
Figure 53 The unprocessed dataset (left) and processed dataset (right) with a Gaussian convolution and scale factor.¶
Applying Operations along a Dimension¶
Multi-dimensional NMR simulations may need different operations applied along different dimensions. Each operation has the attribute dim_index, which is used to apply operations along a certain dimension.
By default, dim_index is None and is applied along the 1st dimension. An integer or list of integers can be passed to dim_index, specifying the dimensions. Below are examples of specifying the dimensions
# Gaussian apodization along the first dimension (default)
sp.apodization.Gaussian(FWHM="10 Hz")
# Constant offset along the second dimension
sp.baseline.ConstantOffset(offset=10, dim_index=1)
# Exponential apodization along the first and third dimensions
sp.apodization.Exponential(FWHM="10 Hz", dim_index=[0, 2])
Applying Apodizations to specific Dependent Variables¶
Each dimension in a simulated spectrum can hold multiple dependent variables (a.k.a. contributions from multiple spin systems). Each spin system may need different convolutions applied to match an experimental spectrum. The
Apodization sub-classes have the dv_index attribute, specifying which dependent variable (spin system) to apply the operation on. By default, dv_index is None and will apply the convolution to all dependent variables
in a dimension.
Note
The index of a dependent variable (spin system) corresponds to the order of spin systems in the
spin_systems list.
processor = sp.SignalProcessor(
operations=[
sp.IFFT(),
sp.apodization.Gaussian(FWHM="25 Hz", dv_index=0),
sp.apodization.Gaussian(FWHM="70 Hz", dv_index=1),
sp.IFFT(),
]
)
The above list of operations will apply 25 and 70 Hz of Gaussian line broadening to dependent variables at index 0 and 1, respectively.
Let’s add another dependent variable to the previously created CSDM object to target specific dependent variables.
test_data = np.zeros(500)
test_data[300] = 1
csdm_object.add_dependent_variable(cp.as_dependent_variable(test_data))
Now, we again apply the operations with the
apply_operations() method.
The comparison of the unprocessed and processed dataset is also shown below.
processed_dataset = processor.apply_operations(dataset = csdm_object)
Below is a plot of the dataset before and after applying the operations
_, ax = plt.subplots(1, 2, figsize=(8, 3), subplot_kw={"projection":"csdm"})
ax[0].plot(csdm_object, linewidth=1)
ax[0].set_title("Unprocessed")
ax[1].plot(processed_dataset.real, linewidth=1)
ax[1].set_title("Processed")
plt.tight_layout()
plt.show()
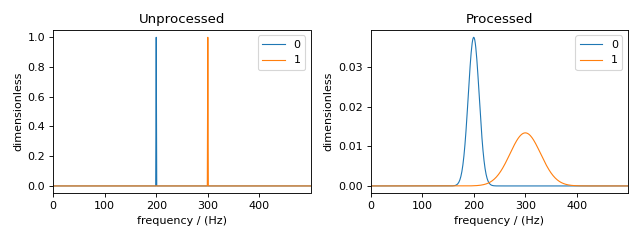
Figure 55 The unprocessed dataset (left) and the processed dataset (right) with convolutions applied to different dependent variables.¶
