Welcome to the Mrsimulator documentation¶
Deployment |


|
Build Status |
|
License |

|
Metrics |
|
Social |




|
About
mrsimulator is an open-source Python package for fast computation/analysis of nuclear magnetic resonance (NMR) spectra in fluid and solid phases.
See our example galleries
Why use mrsimulator?
It is open-source and free.
It is a fast and versatile multi-dimensional solid-state NMR spectra simulator, including MAS and VAS spectra of nuclei experiencing chemical shift (nuclear shielding) and quadrupolar coupling interactions.
It includes simulation of weakly coupled nuclei experiencing J and dipolar couplings.
It is fully documented with a stable and simple API and is easily incorporated into Python scripts and web apps.
It is compatible with modern Python packages, such as Scikit-learn, Keras, etc.
Packages using mrsimulator -
A brief example
from mrsimulator import Simulator, SpinSystem, Site
from mrsimulator.method.lib import BlochDecaySpectrum
import matplotlib.pyplot as plt
# Make Site and SpinSystem objects
H_site = Site(isotope="1H", shielding_symmetric={"zeta": 13.89, "eta": 0.25})
spin_system = SpinSystem(sites=[H_site])
# Make static and MAS one-pulse acquire Method objects
static = BlochDecaySpectrum(channels=["1H"])
mas = BlochDecaySpectrum(channels=["1H"], rotor_frequency=1000) # in Hz
# Setup and run the Simulation object
sim = Simulator(spin_systems=[spin_system], methods=[static, mas])
sim.run()
# Plot the spectra
fig, ax = plt.subplots(1, 2, figsize=(6, 3), subplot_kw={"projection": "csdm"})
ax[0].plot(sim.methods[0].simulation)
ax[0].set_title("Static")
ax[1].plot(sim.methods[1].simulation)
ax[1].set_title("MAS")
plt.tight_layout()
plt.show()
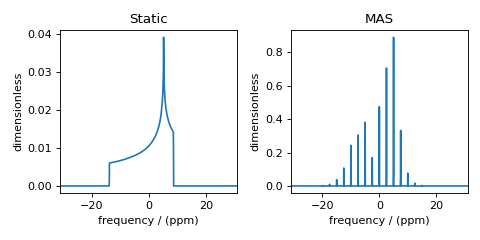
Figure 1 Simulation of static and MAS solid-state NMR spectra¶
Note
Throughout the web version of this documentation, you can copy code blocks into your clipboard by hovering over the top right corner of each gray code block and clicking the copy-to-clipboard icon. This is useful for copying code examples into your Python scripts and Jupyter notebooks.
Features
The mrsimulator package offers the following
Fast simulation of one and two-dimensional solid-state NMR spectra.
- Simulation of coupled and uncoupled spin system
for spin \(I=\frac{1}{2}\), and quadrupole \(I \ge \frac{1}{2}\) nuclei
at arbitrary macroscopic magnetic flux density
at arbitrary rotor angles
at arbitrary spinning frequency
- A library of pre-built NMR methods,
1D Bloch decay spectrum
1D Bloch decay central transition spectrum
2D Multi-Quantum Variable Angle Spinning (MQ-VAS)
2D Satellite-Transition Variable Angle Spinning (ST-VAS)
2D isotropic/anisotropic sideband correlation spectrum (e.g. PASS and MAT)
2D Magic-Angle Flipping (MAF)
2D Dynamic-Angle Spinning (DAS)
Custom user-defined methods (Method)
- Models for tensor parameter distribution in amorphous materials.
Czjzek
Extended Czjzek
Custom user-defined models
Introduction¶
User Guide¶
User Documentation
- Spin System
- Spin System Distributions
- Methods Library
- Method
- Simulator
- Signal Processor
- mrsimulator I/O
Examples¶
Theory¶
API and references¶
API and references
Project details¶
Project details
Reporting Bugs¶
Submit bug reports or feature requests on the Github issue tracker.
Discussions are welcome on the Github discussion page.
How to cite¶
Please refer to mrsimulator Github page for details.
