Signal Processor¶
After running a simulation, you may need to apply some post-simulation signal processing. For example, you may need to scale the simulated spectrum to match experimental intensities, or you may want to convolve the spectrum with a Lorentzian, Gaussian, or other line-broadening
function. For this reason, mrsimulator offers some frequently used NMR signal processing tools within the mrsimulator.signal_processor module.
See also
Signal Processing Gallery for notebooks using common processing functions.
CSDM object¶
The simulated spectrum is held in a CSDM [1] object, which supports multi-dimensional scientific datasets (NMR, EPR, FTIR, GC, etc.). For more information, see the csdmpy documentation.
SignalProcessor class¶
Signal processing is a series of operations sequentially applied to the dataset.
In mrsimulator, the SignalProcessor object is
used to apply operations. Here we create a new SignalProcessor object
# Import the signal_processor module
from mrsimulator import signal_processor as sp
# Create a new SignalProcessor object
processor = sp.SignalProcessor()
Each signal processor object holds a list of operations under the operations attribute. Below we add operations to apply Gaussian line broadening and a scale factor.
processor.operations = [
sp.IFFT(),
sp.apodization.Gaussian(FWHM="50 Hz"),
sp.FFT(),
sp.Scale(factor=120),
]
First, an inverse Fourier transform is applied to the dataset. Then,
a Gaussian apodization with a full-width-at-half-maximum of 50 Hz
in the frequency domain is applied. The unit used for the FWHM
attribute corresponds to the dimensionality of the dataset. By
choosing Hz, we imply the dataset is in units of frequency.
Finally, a forward Fourier transform is applied to the apodized
dataset, and all points are scaled up by 120 times.
Note
Convolutions in mrsimulator are performed using the Convolution Theorem. A spectrum is Fourier transformed, and apodizations are performed in the time domain before being transformed back into the frequency domain.
Let’s create a CSDM object and then apply the operations to visualize the results.
import csdmpy as cp
import numpy as np
# Create a CSDM object with delta function at 200 Hz
test_data = np.zeros(500)
test_data[200] = 1
csdm_object = cp.CSDM(
dependent_variables=[cp.as_dependent_variable(test_data)],
dimensions=[cp.LinearDimension(count=500, increment="1 Hz")],
)
To apply the previously defined signal processing operations to the above CSDM object, use
the apply_operations() method of the
SignalProcessor instance as follows
processed_dataset = processor.apply_operations(dataset = csdm_object)
The variable processed_dataset is another CSDM object holding the dataset
after the list of operations has been applied to csdm_object. Below is a
plot comparing the unprocessed and processed dataset
import matplotlib.pyplot as plt
_, ax = plt.subplots(1, 2, figsize = (8, 3), subplot_kw = {"projection":"csdm"})
ax[0].plot(csdm_object, color="black", linewidth=1)
ax[0].set_title("Unprocessed")
ax[1].plot(processed_dataset.real, color="black", linewidth=1)
ax[1].set_title("Processed")
plt.tight_layout()
plt.show()
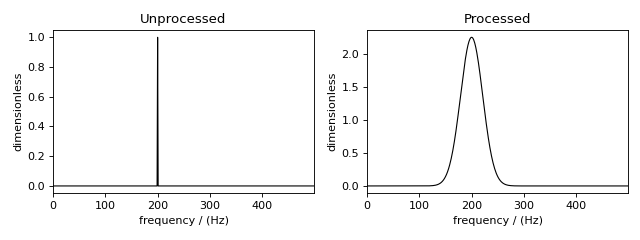
Figure 90 The unprocessed dataset (left) and processed dataset (right) with a Gaussian convolution and scale factor.¶
Applying Operations along a Dimension¶
Multi-dimensional NMR simulations may need different operations applied along different dimensions. Each operation has the attribute dim_index, which is used to apply operations along a certain dimension.
By default, dim_index is None and is applied along the 1st dimension. An integer or list of integers can be passed to dim_index, specifying the dimensions. Below are examples of specifying the dimensions
# Gaussian apodization along the first dimension (default)
sp.apodization.Gaussian(FWHM="10 Hz")
# Constant offset along the second dimension
sp.baseline.ConstantOffset(offset=10, dim_index=1)
# Exponential apodization along the first and third dimensions
sp.apodization.Exponential(FWHM="10 Hz", dim_index=[0, 2])
Applying Apodizations to specific Dependent Variables¶
Each dimension in a simulated spectrum can hold multiple dependent variables (a.k.a. contributions from multiple spin systems). Each spin system may need different convolutions applied to match an experimental spectrum. The
Apodization sub-classes have the dv_index attribute, specifying which dependent variable (spin system) to apply the operation on. By default, dv_index is None and will apply the convolution to all dependent variables
in a dimension.
Note
The index of a dependent variable (spin system) corresponds to the order of spin systems in the
spin_systems list.
processor = sp.SignalProcessor(
operations=[
sp.IFFT(),
sp.apodization.Gaussian(FWHM="25 Hz", dv_index=0),
sp.apodization.Gaussian(FWHM="70 Hz", dv_index=1),
sp.IFFT(),
]
)
The above list of operations will apply 25 and 70 Hz of Gaussian line broadening to dependent variables at index 0 and 1, respectively.
Let’s add another dependent variable to the previously created CSDM object to target specific dependent variables.
test_data = np.zeros(500)
test_data[300] = 1
csdm_object.add_dependent_variable(cp.as_dependent_variable(test_data))
Now, we again apply the operations with the
apply_operations() method.
The comparison of the unprocessed and processed dataset is also shown below.
processed_dataset = processor.apply_operations(dataset = csdm_object)
Below is a plot of the dataset before and after applying the operations
_, ax = plt.subplots(1, 2, figsize=(8, 3), subplot_kw={"projection":"csdm"})
ax[0].plot(csdm_object, linewidth=1)
ax[0].set_title("Unprocessed")
ax[1].plot(processed_dataset.real, linewidth=1)
ax[1].set_title("Processed")
plt.tight_layout()
plt.show()
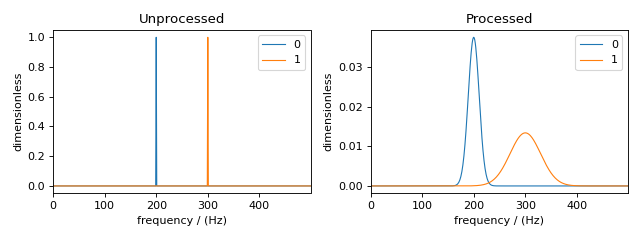
Figure 92 The unprocessed dataset (left) and the processed dataset (right) with convolutions applied to different dependent variables.¶
