Note
Click here to download the full example code
Protein GB1, ¹³C and ¹⁵N (I=1/2)¶
¹³C/¹⁵N (I=1/2) spinning sideband simulation.
The following is the spinning sideband simulation of a macromolecule, protein GB1. The \(^{13}\text{C}\) and \(^{15}\text{N}\) CSA tensor parameters were obtained from Hung et al. [1], which consists of 42 \(^{13}\text{C}\alpha\), 44 \(^{13}\text{CO}\), and 44 \(^{15}\text{NH}\) tensors. In the following example, instead of creating 130 spin systems, we download the spin systems from a remote file and load it directly to the Simulator object.
import matplotlib.pyplot as plt
from mrsimulator import Simulator
from mrsimulator.method.lib import BlochDecaySpectrum
from mrsimulator.method import SpectralDimension
from mrsimulator import signal_processor as sp
Create the Simulator object and load the spin systems from an external file.
sim = Simulator()
host = "https://ssnmr.org/sites/default/files/mrsimulator/"
filename = "protein_GB1_15N_13CA_13CO.mrsys"
sim.load_spin_systems(host + filename) # load the spin systems.
print(f"number of spin systems = {len(sim.spin_systems)}")
Out:
number of spin systems = 130
all_sites = sim.sites().to_pd()
all_sites.head()
Create a \(^{13}\text{C}\) Bloch decay spectrum method.
method_13C = BlochDecaySpectrum(
channels=["13C"],
magnetic_flux_density=11.74, # in T
rotor_frequency=3000, # in Hz
spectral_dimensions=[
SpectralDimension(
count=8192,
spectral_width=5e4, # in Hz
reference_offset=2e4, # in Hz
label=r"$^{13}$C resonances",
)
],
)
Since the spin systems contain both \(^{13}\text{C}\) and \(^{15}\text{N}\) sites, let’s also create a \(^{15}\text{N}\) Bloch decay spectrum method.
method_15N = BlochDecaySpectrum(
channels=["15N"],
magnetic_flux_density=11.74, # in T
rotor_frequency=3000, # in Hz
spectral_dimensions=[
SpectralDimension(
count=8192,
spectral_width=4e4, # in Hz
reference_offset=7e3, # in Hz
label=r"$^{15}$N resonances",
)
],
)
Add the methods to the Simulator object and run the simulation
# Add the methods.
sim.methods = [method_13C, method_15N]
# Run the simulation.
sim.run()
# Get the simulation dataset from the respective methods.
dataset_13C = sim.methods[0].simulation # method at index 0 is 13C Bloch decay method.
dataset_15N = sim.methods[1].simulation # method at index 1 is 15N Bloch decay method.
Add post-simulation signal processing.
processor = sp.SignalProcessor(
operations=[sp.IFFT(), sp.apodization.Exponential(FWHM="10 Hz"), sp.FFT()]
)
# apply post-simulation processing to dataset_13C
processed_dataset_13C = processor.apply_operations(dataset=dataset_13C).real
# apply post-simulation processing to dataset_15N
processed_dataset_15N = processor.apply_operations(dataset=dataset_15N).real
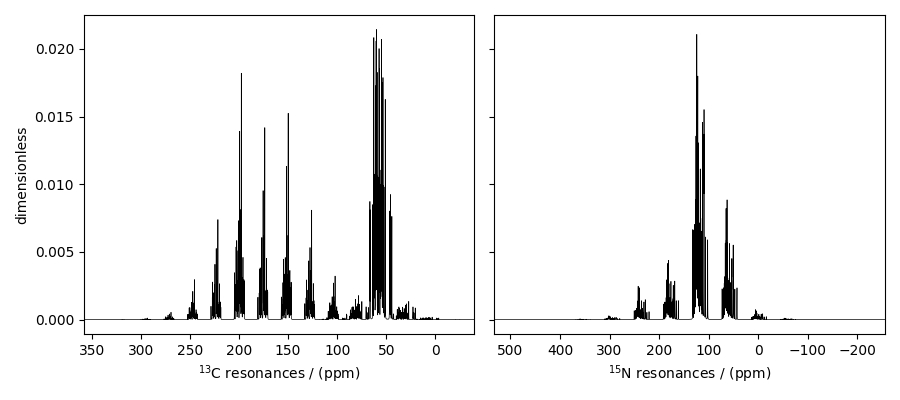
The plot of the simulation after signal processing.
fig, ax = plt.subplots(
1, 2, subplot_kw={"projection": "csdm"}, sharey=True, figsize=(9, 4)
)
ax[0].plot(processed_dataset_13C, color="black", linewidth=0.5)
ax[0].invert_xaxis()
ax[1].plot(processed_dataset_15N, color="black", linewidth=0.5)
ax[1].set_ylabel(None)
ax[1].invert_xaxis()
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 1.890 seconds)
