Note
Click here to download the full example code
¹¹⁹Sn MAS NMR of SnO¶
The following is a spinning sideband manifold fitting example for the 119Sn MAS NMR of SnO. The dataset was acquired and shared by Altenhof et al. [1].
import csdmpy as cp
import matplotlib.pyplot as plt
from lmfit import Minimizer
from mrsimulator import Simulator, SpinSystem, Site, Coupling
from mrsimulator.method.lib import BlochDecaySpectrum
from mrsimulator import signal_processor as sp
from mrsimulator.utils import spectral_fitting as sf
from mrsimulator.utils import get_spectral_dimensions
from mrsimulator.spin_system.tensors import SymmetricTensor
Import the dataset¶
filename = "https://ssnmr.org/sites/default/files/mrsimulator/119Sn_SnO.csdf"
experiment = cp.load(filename)
# standard deviation of noise from the dataset
sigma = 0.6410905
# For spectral fitting, we only focus on the real part of the complex dataset
experiment = experiment.real
# Convert the coordinates along each dimension from Hz to ppm.
_ = [item.to("ppm", "nmr_frequency_ratio") for item in experiment.dimensions]
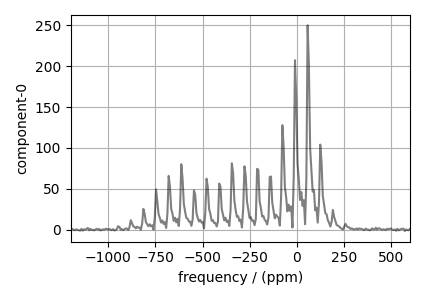
# plot of the dataset.
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
ax.plot(experiment, "k", alpha=0.5)
ax.set_xlim(-1200, 600)
plt.grid()
plt.tight_layout()
plt.show()
Create a fitting model¶
Guess model
Create a guess list of spin systems. There are two spin systems present in this example, - 1) an uncoupled \(^{119}\text{Sn}\) and - 2) a coupled \(^{119}\text{Sn}\)-\(^{117}\text{Sn}\) spin systems.
sn119 = Site(
isotope="119Sn",
isotropic_chemical_shift=-210,
shielding_symmetric=SymmetricTensor(zeta=700, eta=0.1),
)
sn117 = Site(
isotope="117Sn",
isotropic_chemical_shift=0,
)
j_sn = Coupling(
site_index=[0, 1],
isotropic_j=8150.0,
)
sn117_abundance = 7.68 # in %
spin_systems = [
# uncoupled spin system
SpinSystem(sites=[sn119], abundance=100 - sn117_abundance),
# coupled spin systems
SpinSystem(sites=[sn119, sn117], couplings=[j_sn], abundance=sn117_abundance),
]
Method
# Get the spectral dimension parameters from the experiment.
spectral_dims = get_spectral_dimensions(experiment)
MAS = BlochDecaySpectrum(
channels=["119Sn"],
magnetic_flux_density=9.395, # in T
rotor_frequency=10000, # in Hz
spectral_dimensions=spectral_dims,
experiment=experiment, # add the measurement to the method.
)
# Optimize the script by pre-setting the transition pathways for each spin system from
# the method.
for sys in spin_systems:
sys.transition_pathways = MAS.get_transition_pathways(sys)
Guess Spectrum
# Simulation
# ----------
sim = Simulator(spin_systems=spin_systems, methods=[MAS])
sim.run()
# Post Simulation Processing
# --------------------------
processor = sp.SignalProcessor(
operations=[
sp.IFFT(),
sp.apodization.Exponential(FWHM="1500 Hz"),
sp.FFT(),
sp.Scale(factor=5000),
]
)
processed_dataset = processor.apply_operations(dataset=sim.methods[0].simulation).real
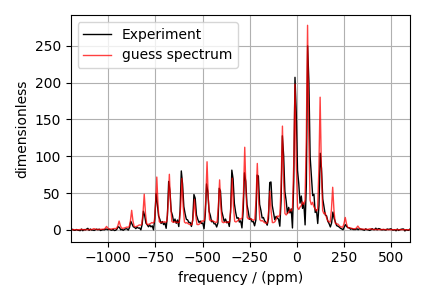
# Plot of the guess Spectrum
# --------------------------
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
ax.plot(experiment, "k", linewidth=1, label="Experiment")
ax.plot(processed_dataset, "r", alpha=0.75, linewidth=1, label="guess spectrum")
ax.set_xlim(-1200, 600)
plt.grid()
plt.legend()
plt.tight_layout()
plt.show()
Least-squares minimization with LMFIT¶
Use the make_LMFIT_params() for a quick
setup of the fitting parameters.
params = sf.make_LMFIT_params(sim, processor, include={"rotor_frequency"})
# Remove the abundance parameters from params. Since the measurement detects 119Sn, we
# also remove the isotropic chemical shift parameter of 117Sn site from params. The
# 117Sn is the site at index 1 of the spin system at index 1.
params.pop("sys_0_abundance")
params.pop("sys_1_abundance")
params.pop("sys_1_site_1_isotropic_chemical_shift")
# Since the 119Sn site is shared between the two spin systems, we add constraints to the
# 119Sn site parameters from the spin system at index 1 to be the same as 119Sn site
# parameters from the spin system at index 0.
lst = [
"isotropic_chemical_shift",
"shielding_symmetric_zeta",
"shielding_symmetric_eta",
]
for item in lst:
params[f"sys_1_site_0_{item}"].expr = f"sys_0_site_0_{item}"
print(params.pretty_print(columns=["value", "min", "max", "vary", "expr"]))
Out:
Name Value Min Max Vary Expr
SP_0_operation_1_Exponential_FWHM 1500 -inf inf True None
SP_0_operation_3_Scale_factor 5000 -inf inf True None
mth_0_rotor_frequency 1e+04 9900 1.01e+04 True None
sys_0_site_0_isotropic_chemical_shift -210 -inf inf True None
sys_0_site_0_shielding_symmetric_eta 0.1 0 1 True None
sys_0_site_0_shielding_symmetric_zeta 700 -inf inf True None
sys_1_coupling_0_isotropic_j 8150 -inf inf True None
sys_1_site_0_isotropic_chemical_shift -210 -inf inf False sys_0_site_0_isotropic_chemical_shift
sys_1_site_0_shielding_symmetric_eta 0.1 0 1 False sys_0_site_0_shielding_symmetric_eta
sys_1_site_0_shielding_symmetric_zeta 700 -inf inf False sys_0_site_0_shielding_symmetric_zeta
None
Solve the minimizer using LMFIT
minner = Minimizer(sf.LMFIT_min_function, params, fcn_args=(sim, processor, sigma))
result = minner.minimize()
result
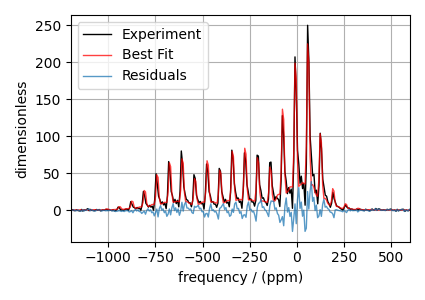
The best fit solution¶
best_fit = sf.bestfit(sim, processor)[0].real
residuals = sf.residuals(sim, processor)[0].real
# Plot the spectrum
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
ax.plot(experiment, "k", linewidth=1, label="Experiment")
ax.plot(best_fit, "r", alpha=0.75, linewidth=1, label="Best Fit")
ax.plot(residuals, alpha=0.75, linewidth=1, label="Residuals")
ax.set_xlim(-1200, 600)
plt.grid()
plt.legend()
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 2.354 seconds)
