Note
Go to the end to download the full example code
Wollastonite, ²⁹Si (I=1/2), MAF¶
²⁹Si (I=1/2) magic angle flipping.
Wollastonite is a high-temperature calcium-silicate, \(\beta−\text{Ca}_3\text{Si}_3\text{O}_9\), with three distinct \(^{29}\text{Si}\) sites. The \(^{29}\text{Si}\) tensor parameters were obtained from Hansen et al. [1]
import numpy as np
import matplotlib.pyplot as plt
from mrsimulator import Simulator, SpinSystem, Site
from mrsimulator import signal_processor as sp
from mrsimulator.spin_system.tensors import SymmetricTensor
from mrsimulator.method import Method, SpectralDimension, SpectralEvent, MixingEvent
Create the sites and spin systems
sites = [
Site(
isotope="29Si",
isotropic_chemical_shift=-89.0, # in ppm
shielding_symmetric=SymmetricTensor(zeta=59.8, eta=0.62), # zeta in ppm
),
Site(
isotope="29Si",
isotropic_chemical_shift=-89.5, # in ppm
shielding_symmetric=SymmetricTensor(zeta=52.1, eta=0.68), # zeta in ppm
),
Site(
isotope="29Si",
isotropic_chemical_shift=-87.8, # in ppm
shielding_symmetric=SymmetricTensor(zeta=69.4, eta=0.60), # zeta in ppm
),
]
spin_systems = [SpinSystem(sites=[s]) for s in sites]
Use the generic Method class to simulate a 2D magic-angle Flipping (MAF) spectrum by customizing the method parameters, as shown below.
Here, we include a MixingEvent with a NoMixing query. A no mixing query
instructs the MAF method to not mix the transitions from the first and second
SpectralEvent. A no mixing query is equivalent to a rotation query where each
channel has a zero phase and angle. Since all spin systems in this example have a
single site, defining no mixing between the two spectral events is superfluous.
We include it such that the method is applicable with multi-site spin systems.
maf = Method(
name="Magic Angle Flipping",
channels=["29Si"],
magnetic_flux_density=14.1, # in T
rotor_frequency=np.inf,
spectral_dimensions=[
SpectralDimension(
count=128,
spectral_width=2e4, # in Hz
label="Anisotropic dimension",
events=[
SpectralEvent(
rotor_angle=90 * np.pi / 180, # in rads
transition_queries=[{"ch1": {"P": [-1], "D": [0]}}],
),
MixingEvent(query="NoMixing"),
],
),
SpectralDimension(
count=128,
spectral_width=3e3, # in Hz
reference_offset=-1.05e4, # in Hz
label="Isotropic dimension",
events=[
SpectralEvent(
rotor_angle=54.735 * np.pi / 180, # in rads
transition_queries=[{"ch1": {"P": [-1], "D": [0]}}],
)
],
),
],
affine_matrix=[[1, -1], [0, 1]],
)
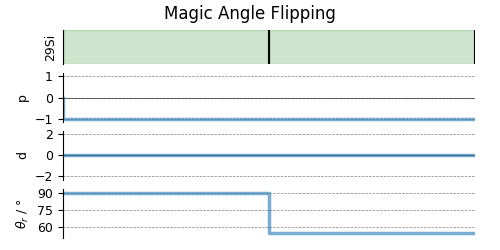
# A graphical representation of the method object.
plt.figure(figsize=(5, 2.5))
maf.plot()
plt.show()
Create the Simulator object, add the method and spin system objects, and run the simulation.
Add post-simulation signal processing.
csdm_dataset = sim.methods[0].simulation
processor = sp.SignalProcessor(
operations=[
sp.IFFT(dim_index=(0, 1)),
sp.apodization.Gaussian(FWHM="50 Hz", dim_index=0),
sp.apodization.Gaussian(FWHM="50 Hz", dim_index=1),
sp.FFT(dim_index=(0, 1)),
]
)
processed_dataset = processor.apply_operations(dataset=csdm_dataset).real
processed_dataset /= processed_dataset.max()
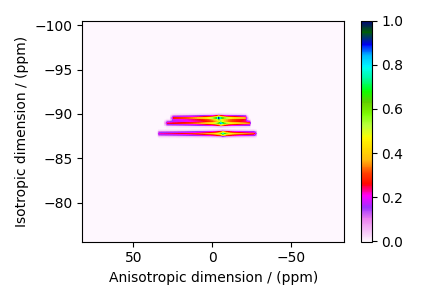
The plot of the simulation after signal processing.
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
cb = ax.imshow(processed_dataset.T, aspect="auto", cmap="gist_ncar_r")
plt.colorbar(cb)
ax.invert_xaxis()
ax.invert_yaxis()
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 0.648 seconds)
