Note
Click here to download the full example code
Coesite, ¹⁷O (I=5/2) DAS¶
¹⁷O (I=5/2) Dynamic-angle spinning (DAS) simulation.
The following is a Dynamic Angle Spinning (DAS) simulation of Coesite. Coesite has five crystallographic \(^{17}\text{O}\) sites. In the following, we use the \(^{17}\text{O}\) EFG tensor information from Grandinetti et al. [1]
import matplotlib.pyplot as plt
import numpy as np
from mrsimulator import Simulator
from mrsimulator import signal_processor as sp
from mrsimulator.method import Method, SpectralDimension, SpectralEvent
Create the Simulator object and load the spin systems database or url address.
sim = Simulator()
# load the spin systems from url.
filename = "https://ssnmr.org/sites/default/files/mrsimulator/coesite_0.mrsys"
sim.load_spin_systems(filename)
Use the generic method, Method, to simulate a 2D DAS spectrum by customizing the method parameters, as shown below.
das = Method(
name="Dynamic Angle Spinning",
channels=["17O"],
magnetic_flux_density=11.74, # in T
rotor_frequency=np.inf,
spectral_dimensions=[
SpectralDimension(
count=256,
spectral_width=5e3, # in Hz
reference_offset=0, # in Hz
label="DAS isotropic dimension",
events=[
SpectralEvent(
fraction=0.5,
rotor_angle=37.38 * 3.14159 / 180, # in rads
transition_queries=[{"ch1": {"P": [-1], "D": [0]}}],
),
SpectralEvent(
fraction=0.5,
rotor_angle=79.19 * 3.14159 / 180, # in rads
transition_queries=[{"ch1": {"P": [-1], "D": [0]}}],
),
],
),
# The last spectral dimension block is the direct-dimension
SpectralDimension(
count=256,
spectral_width=2e4, # in Hz
reference_offset=0, # in Hz
label="MAS dimension",
events=[
SpectralEvent(
rotor_angle=54.735 * 3.14159 / 180, # in rads
transition_queries=[{"ch1": {"P": [-1], "D": [0]}}],
)
],
),
],
)
sim.methods = [das] # add the method
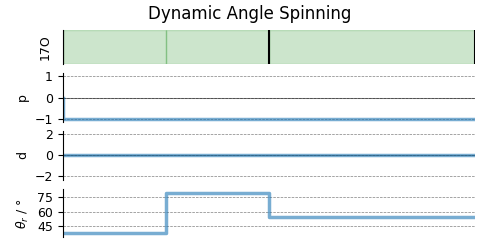
# A graphical representation of the method object.
plt.figure(figsize=(5, 2.5))
das.plot()
plt.show()
Run the simulation
sim.run()
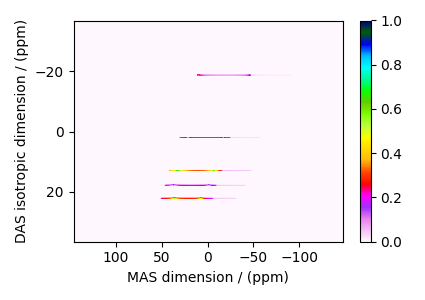
The plot of the simulation.
dataset = sim.methods[0].simulation
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
cb = ax.imshow(dataset.real / dataset.real.max(), aspect="auto", cmap="gist_ncar_r")
plt.colorbar(cb)
ax.invert_xaxis()
ax.invert_yaxis()
plt.tight_layout()
plt.show()
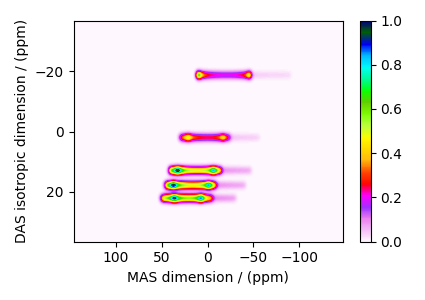
Add post-simulation signal processing.
processor = sp.SignalProcessor(
operations=[
# Gaussian convolution along both dimensions.
sp.IFFT(dim_index=(0, 1)),
sp.apodization.Gaussian(FWHM="0.3 kHz", dim_index=0),
sp.apodization.Gaussian(FWHM="0.15 kHz", dim_index=1),
sp.FFT(dim_index=(0, 1)),
]
)
processed_dataset = processor.apply_operations(dataset=dataset)
processed_dataset /= processed_dataset.max()
The plot of the simulation after signal processing.
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
cb = ax.imshow(processed_dataset.real, cmap="gist_ncar_r", aspect="auto")
plt.colorbar(cb)
ax.invert_xaxis()
ax.invert_yaxis()
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 1.024 seconds)
