Note
Click here to download the full example code
Wollastonite, ²⁹Si (I=1/2), MAF¶
²⁹Si (I=1/2) magic angle flipping.
Wollastonite is a high-temperature calcium-silicate, \(\beta−\text{Ca}_3\text{Si}_3\text{O}_9\), with three distinct \(^{29}\text{Si}\) sites. The \(^{29}\text{Si}\) tensor parameters were obtained from Hansen et al. [1]
import matplotlib.pyplot as plt
import numpy as np
from mrsimulator import Simulator, SpinSystem, Site
from mrsimulator import signal_processor as sp
from mrsimulator.spin_system.tensors import SymmetricTensor
from mrsimulator.method import Method, SpectralDimension, SpectralEvent, MixingEvent
Create the sites and spin systems
sites = [
Site(
isotope="29Si",
isotropic_chemical_shift=-89.0, # in ppm
shielding_symmetric=SymmetricTensor(zeta=59.8, eta=0.62), # zeta in ppm
),
Site(
isotope="29Si",
isotropic_chemical_shift=-89.5, # in ppm
shielding_symmetric=SymmetricTensor(zeta=52.1, eta=0.68), # zeta in ppm
),
Site(
isotope="29Si",
isotropic_chemical_shift=-87.8, # in ppm
shielding_symmetric=SymmetricTensor(zeta=69.4, eta=0.60), # zeta in ppm
),
]
spin_systems = [SpinSystem(sites=[s]) for s in sites]
Use the generic method, Method, to simulate a 2D Magic-Angle Flipping (MAF) spectrum by customizing the method parameters, as shown below.
Here we include the special MixingEvent with query NoMixing to tell the MAF
method to not connect any of the transitions between the first and second
SpectralEvent. A query of NoMixing is equivalent to a rotational query where
each channel has a phase and angle of 0. Since all spin systems in this example have
a single site, defining no mixing between the two spectral events is superfluous, but
we include it so this method may be used with multi-site spin systems.
maf = Method(
name="Magic Angle Flipping",
channels=["29Si"],
magnetic_flux_density=14.1, # in T
rotor_frequency=np.inf,
spectral_dimensions=[
SpectralDimension(
count=128,
spectral_width=2e4, # in Hz
label="Anisotropic dimension",
events=[
SpectralEvent(
rotor_angle=90 * 3.14159 / 180, # in rads
transition_queries=[{"ch1": {"P": [-1], "D": [0]}}],
),
MixingEvent(query="NoMixing"),
],
),
SpectralDimension(
count=128,
spectral_width=3e3, # in Hz
reference_offset=-1.05e4, # in Hz
label="Isotropic dimension",
events=[
SpectralEvent(
rotor_angle=54.735 * 3.14159 / 180, # in rads
transition_queries=[{"ch1": {"P": [-1], "D": [0]}}],
)
],
),
],
affine_matrix=[[1, -1], [0, 1]],
)
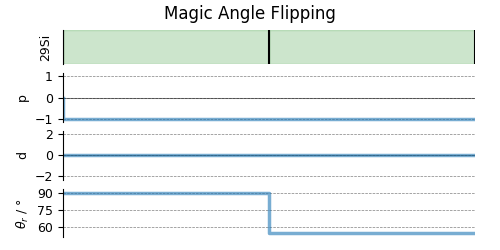
# A graphical representation of the method object.
plt.figure(figsize=(5, 2.5))
maf.plot()
plt.show()
Create the Simulator object, add the method and spin system objects, and run the simulation.
Add post-simulation signal processing.
csdm_dataset = sim.methods[0].simulation
processor = sp.SignalProcessor(
operations=[
sp.IFFT(dim_index=(0, 1)),
sp.apodization.Gaussian(FWHM="50 Hz", dim_index=0),
sp.apodization.Gaussian(FWHM="50 Hz", dim_index=1),
sp.FFT(dim_index=(0, 1)),
]
)
processed_dataset = processor.apply_operations(dataset=csdm_dataset).real
processed_dataset /= processed_dataset.max()
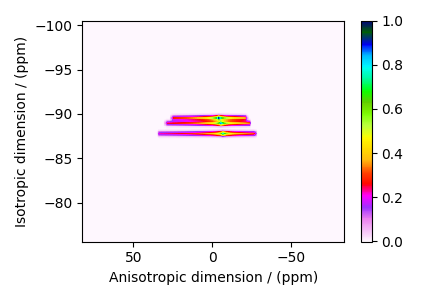
The plot of the simulation after signal processing.
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
cb = ax.imshow(processed_dataset.T, aspect="auto", cmap="gist_ncar_r")
plt.colorbar(cb)
ax.invert_xaxis()
ax.invert_yaxis()
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 0.623 seconds)
