Note
Click here to download the full example code
MCl₂.2D₂O, ²H (I=1) Shifting-d echo¶
²H (I=1) 2D NMR CSA-Quad 1st order correlation spectrum simulation.
The following is a static shifting-d echo NMR correlation simulation of \(\text{MCl}_2\cdot 2\text{D}_2\text{O}\) crystalline solid, where \(M \in [\text{Cu}, \text{Ni}, \text{Co}, \text{Fe}, \text{Mn}]\). The tensor parameters for the simulation and the corresponding spectrum are reported by Walder et al. [1].
import matplotlib.pyplot as plt
from mrsimulator import Simulator, SpinSystem, Site
from mrsimulator import signal_processor as sp
from mrsimulator.spin_system.tensors import SymmetricTensor
from mrsimulator.method import Method, SpectralDimension, SpectralEvent, MixingEvent
Generate the site and spin system objects.
site_Ni = Site(
isotope="2H",
isotropic_chemical_shift=-97, # in ppm
shielding_symmetric=SymmetricTensor(
zeta=-551, # in ppm
eta=0.12,
alpha=62 * 3.14159 / 180, # in rads
beta=114 * 3.14159 / 180, # in rads
gamma=171 * 3.14159 / 180, # in rads
),
quadrupolar=SymmetricTensor(Cq=77.2e3, eta=0.9), # Cq in Hz
)
site_Cu = Site(
isotope="2H",
isotropic_chemical_shift=51, # in ppm
shielding_symmetric=SymmetricTensor(
zeta=146, # in ppm
eta=0.84,
alpha=95 * 3.14159 / 180, # in rads
beta=90 * 3.14159 / 180, # in rads
gamma=0 * 3.14159 / 180, # in rads
),
quadrupolar=SymmetricTensor(Cq=118.2e3, eta=0.8), # Cq in Hz
)
site_Co = Site(
isotope="2H",
isotropic_chemical_shift=215, # in ppm
shielding_symmetric=SymmetricTensor(
zeta=-1310, # in ppm
eta=0.23,
alpha=180 * 3.14159 / 180, # in rads
beta=90 * 3.14159 / 180, # in rads
gamma=90 * 3.14159 / 180, # in rads
),
quadrupolar=SymmetricTensor(Cq=114.6e3, eta=0.95), # Cq in Hz
)
site_Fe = Site(
isotope="2H",
isotropic_chemical_shift=101, # in ppm
shielding_symmetric=SymmetricTensor(
zeta=-1187, # in ppm
eta=0.4,
alpha=122 * 3.14159 / 180, # in rads
beta=90 * 3.14159 / 180, # in rads
gamma=90 * 3.14159 / 180, # in rads
),
quadrupolar=SymmetricTensor(Cq=114.2e3, eta=0.98), # Cq in Hz
)
site_Mn = Site(
isotope="2H",
isotropic_chemical_shift=145, # in ppm
shielding_symmetric=SymmetricTensor(
zeta=-1236, # in ppm
eta=0.23,
alpha=136 * 3.14159 / 180, # in rads
beta=90 * 3.14159 / 180, # in rads
gamma=90 * 3.14159 / 180, # in rads
),
quadrupolar=SymmetricTensor(Cq=1.114e5, eta=1.0), # Cq in Hz
)
spin_systems = [
SpinSystem(sites=[s], name=f"{n}Cl$_2$.2D$_2$O")
for s, n in zip(
[site_Ni, site_Cu, site_Co, site_Fe, site_Mn], ["Ni", "Cu", "Co", "Fe", "Mn"]
)
]
Use the generic method, Method, to generate a 2D shifting-d echo method. The
reported shifting-d 2D sequence is a correlation of the shielding frequencies to the
first-order quadrupolar frequencies. Here, we create a correlation method using the
freq_contrib attribute, which acts as a switch
for including the frequency contributions from interaction during the event.
In the following method, we assign the ["Quad1_2"] and
["Shielding1_0", "Shielding1_2"] as the value to the freq_contrib key. The
Quad1_2 is an enumeration for selecting the first-order second-rank quadrupolar
frequency contributions. Shielding1_0 and Shielding1_2 are enumerations for
the first-order shielding with zeroth and second-rank tensor contributions,
respectively. See FrequencyEnum for details.
Like the previous example, we stipulate no mixing between the two spectral events
using a MixingEvent with NoMixing as the query. Since all spin systems in this
example have a single site, defining no mixing between the two spectral events is
superfluous, but we include it so this method may be used with multi-site spin
systems.
shifting_d = Method(
name="Shifting-d",
channels=["2H"],
magnetic_flux_density=9.395, # in T
rotor_frequency=0, # in Hz
spectral_dimensions=[
SpectralDimension(
count=512,
spectral_width=2.5e5, # in Hz
label="Quadrupolar frequency",
events=[
SpectralEvent(
transition_queries=[{"ch1": {"P": [-1]}}],
freq_contrib=["Quad1_2"],
),
MixingEvent(query="NoMixing"),
],
),
SpectralDimension(
count=256,
spectral_width=2e5, # in Hz
reference_offset=2e4, # in Hz
label="Paramagnetic shift",
events=[
SpectralEvent(
transition_queries=[{"ch1": {"P": [-1]}}],
freq_contrib=["Shielding1_0", "Shielding1_2"],
)
],
),
],
)
# A graphical representation of the method object.
plt.figure(figsize=(4, 1.5))
shifting_d.plot()
plt.show()
Create the Simulator object, add the method and spin system objects, and run the simulation.
sim = Simulator(spin_systems=spin_systems, methods=[shifting_d])
# Configure the simulator object. For non-coincidental tensors, set the value of the
# `integration_volume` attribute to `hemisphere`.
sim.config.integration_volume = "hemisphere"
sim.config.decompose_spectrum = "spin_system" # simulate spectra per spin system
sim.run()
Add post-simulation signal processing.
dataset = sim.methods[0].simulation
processor = sp.SignalProcessor(
operations=[
# Gaussian convolution along both dimensions.
sp.IFFT(dim_index=(0, 1)),
sp.apodization.Gaussian(FWHM="9 kHz", dim_index=0), # along dimension 0
sp.apodization.Gaussian(FWHM="9 kHz", dim_index=1), # along dimension 1
sp.FFT(dim_index=(0, 1)),
]
)
processed_dataset = processor.apply_operations(dataset=dataset)
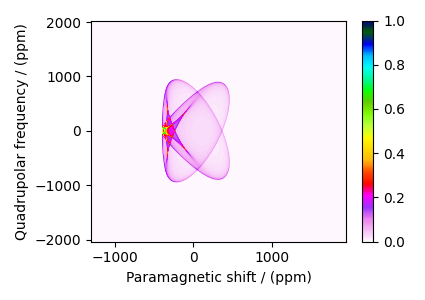
The plot of the simulation. Because we configured the simulator object to simulate spectrum per spin system, the following dataset is a CSDM object containing five simulations (dependent variables). Let’s visualize the first dataset corresponding to \(\text{NiCl}_2\cdot 2 \text{D}_2\text{O}\).
dataset_Ni = dataset.split()[0].real
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
cb = ax.imshow(dataset_Ni / dataset_Ni.max(), aspect="auto", cmap="gist_ncar_r")
plt.title(None)
plt.colorbar(cb)
plt.tight_layout()
plt.show()
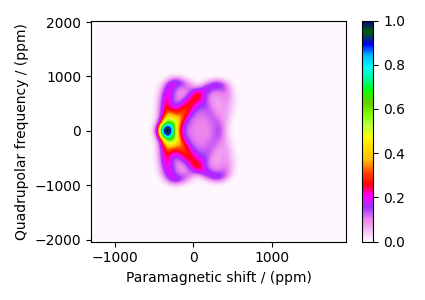
The plot of the simulation after signal processing.
proc_dataset_Ni = processed_dataset.split()[0].real
plt.figure(figsize=(4.25, 3.0))
ax = plt.subplot(projection="csdm")
cb = ax.imshow(
proc_dataset_Ni / proc_dataset_Ni.max(), cmap="gist_ncar_r", aspect="auto"
)
plt.title(None)
plt.colorbar(cb)
plt.tight_layout()
plt.show()
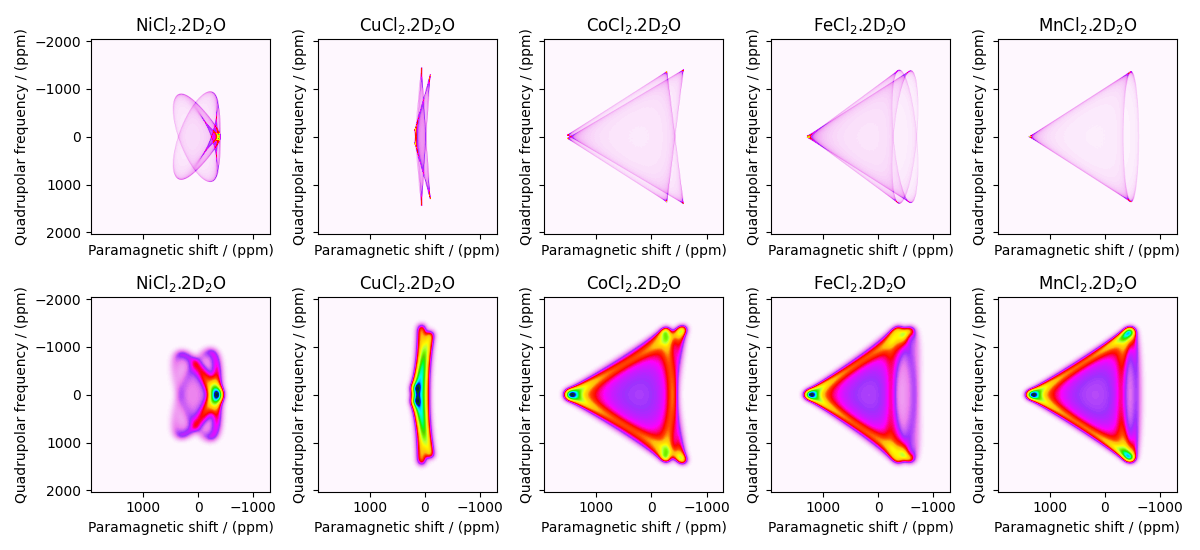
Let’s plot all the simulated datasets.
fig, ax = plt.subplots(
2, 5, sharex=True, sharey=True, figsize=(12, 5.5), subplot_kw={"projection": "csdm"}
)
for i, dataset_obj in enumerate([dataset, processed_dataset]):
for j, datum in enumerate(dataset_obj.split()):
ax[i, j].imshow((datum / datum.max()).real, aspect="auto", cmap="gist_ncar_r")
ax[i, j].invert_xaxis()
ax[i, j].invert_yaxis()
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 2.549 seconds)
