Note
Click here to download the full example code
¹³C MAS NMR of Glycine (CSA) multi-spectra fit¶
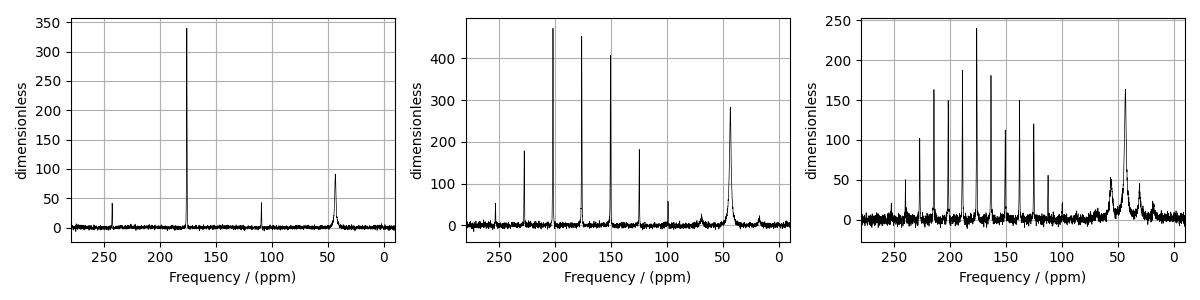
The following is a multi-dataset least-squares fitting example of \(^{13}\text{C}\) MAS NMR spectrum of Glycine spinning at 5 kHz, 1.94 kHz, and 960 Hz. Before trying multi-dataset fitting, we recommend that you first try individual fits. The experimental datasets are part of DMFIT [1] examples.
import csdmpy as cp
import matplotlib.pyplot as plt
from lmfit import Minimizer
from mrsimulator import Simulator, SpinSystem, Site
from mrsimulator.method.lib import BlochDecaySpectrum
from mrsimulator import signal_processor as sp
from mrsimulator.utils import spectral_fitting as sf
from mrsimulator.utils import get_spectral_dimensions
from mrsimulator.spin_system.tensors import SymmetricTensor
Import the datasets¶
Import the datasets and assign the standard deviation of noise for each dataset. Here,
sigma1, sigma2, and sigma3 are the noise standard deviation for the
dataset acquired at 5 kHz, 1.94 kHz, and 960 Hz spinning speeds, respectively.
host = "https://nmr.cemhti.cnrs-orleans.fr/Dmfit/Help/csdm/"
filename1 = "13C MAS 5000Hz - Glycine.csdf"
filename2 = "13C MAS 1940Hz - Glycine.csdf"
filename3 = "13C MAS 960Hz - Glycine.csdf"
experiment1 = cp.load(host + filename1).real
experiment2 = cp.load(host + filename2).real
experiment3 = cp.load(host + filename3).real
experiments = [experiment1, experiment2, experiment3]
# standard deviation of noise from the dataset
sigma1 = 1.97637
sigma2 = 3.822249
sigma3 = 3.982936
sigmas = [sigma1, sigma2, sigma3]
# Convert the coordinates along each dimension from Hz to ppm for each dataset
fig, ax = plt.subplots(1, 3, figsize=(12, 3), subplot_kw={"projection": "csdm"})
for i, experiment in enumerate(experiments):
_ = [item.to("ppm", "nmr_frequency_ratio") for item in experiment.dimensions]
# plot of the dataset.
ax[i].plot(experiment, color="black", linewidth=0.5, label="Experiment")
ax[i].set_xlim(280, -10)
ax[i].grid()
plt.tight_layout()
plt.show()
Create a fitting model¶
Spin System: The objective of a multi-dataset fitting is to optimize the spin system parameters using multiple datasets. In this example, we create two single-site spin systems, which are then shared by three method objects.
C1 = Site(
isotope="13C",
isotropic_chemical_shift=176.0, # in ppm
shielding_symmetric=SymmetricTensor(zeta=60, eta=0.6), # zeta in Hz
)
C2 = Site(
isotope="13C",
isotropic_chemical_shift=43.0, # in ppm
shielding_symmetric=SymmetricTensor(zeta=30, eta=0.5), # zeta in Hz
)
spin_systems = [SpinSystem(sites=[C1], name="C1"), SpinSystem(sites=[C2], name="C2")]
Method: Create the three MAS method objects with respective MAS spinning speeds.
# Get the spectral dimension parameters from the respective experiment and setup the
# corresponding method.
# Method for dataset 1
spectral_dims1 = get_spectral_dimensions(experiment1)
MAS1 = BlochDecaySpectrum(
channels=["13C"],
magnetic_flux_density=7.05, # in T
rotor_frequency=5000, # in Hz
spectral_dimensions=spectral_dims1,
experiment=experiment1, # add experimental dataset 1
)
# Method for dataset 2
spectral_dims2 = get_spectral_dimensions(experiment2)
MAS2 = BlochDecaySpectrum(
channels=["13C"],
magnetic_flux_density=7.05, # in T
rotor_frequency=1940, # in Hz
spectral_dimensions=spectral_dims2,
experiment=experiment2, # add experimental dataset 2
)
# Method for dataset 3
spectral_dims3 = get_spectral_dimensions(experiment3)
MAS3 = BlochDecaySpectrum(
channels=["13C"],
magnetic_flux_density=7.05, # in T
rotor_frequency=960, # in Hz
spectral_dimensions=spectral_dims3,
experiment=experiment3, # add experimental dataset 3
)
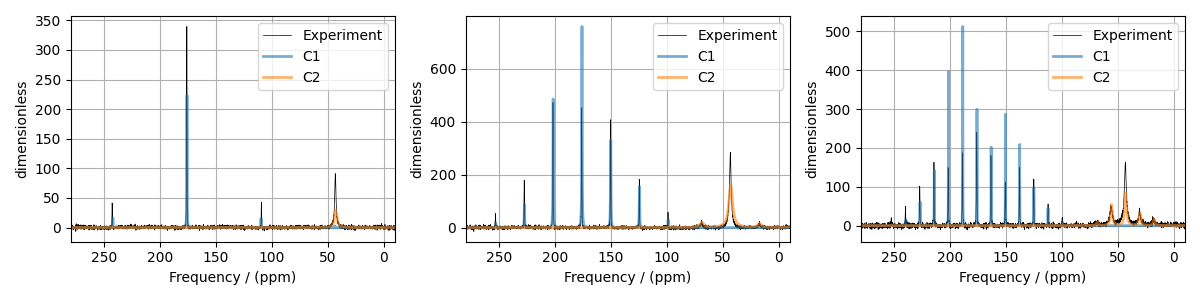
Guess Model Spectrum
# Simulation
# ----------
# Add the spin systems and the three methods to the simulator object.
sim = Simulator(spin_systems=spin_systems, methods=[MAS1, MAS2, MAS3])
sim.config.decompose_spectrum = "spin_system"
sim.run()
# Post Simulation Processing
# --------------------------
# Add signal processing to simulation dataset from the three methods.
# Processor for dataset 1
processor1 = sp.SignalProcessor(
operations=[
sp.IFFT(),
sp.apodization.Exponential(FWHM="20 Hz", dv_index=0), # spin system 0
sp.apodization.Exponential(FWHM="200 Hz", dv_index=1), # spin system 1
sp.FFT(),
sp.Scale(factor=10), # dataset is scaled independently using scale factor.
]
)
# Processor for dataset 2
processor2 = sp.SignalProcessor(
operations=[
sp.IFFT(),
sp.apodization.Exponential(FWHM="30 Hz", dv_index=0), # spin system 0
sp.apodization.Exponential(FWHM="300 Hz", dv_index=1), # spin system 1
sp.FFT(),
sp.Scale(factor=100), # dataset is scaled independently using scale factor.
]
)
# Processor for dataset 3
processor3 = sp.SignalProcessor(
operations=[
sp.IFFT(),
sp.apodization.Exponential(FWHM="10 Hz", dv_index=0), # spin system 0
sp.apodization.Exponential(FWHM="150 Hz", dv_index=1), # spin system 1
sp.FFT(),
sp.Scale(factor=50), # dataset is scaled independently using scale factor.
]
)
processors = [processor1, processor2, processor3]
processed_dataset = []
for i, proc in enumerate(processors):
processed_dataset.append(
proc.apply_operations(dataset=sim.methods[i].simulation).real
)
# Plot of the guess Spectrum
# --------------------------
fig, ax = plt.subplots(1, 3, figsize=(12, 3), subplot_kw={"projection": "csdm"})
for i, exp_dataset in enumerate(experiments):
ax[i].plot(exp_dataset, color="black", linewidth=0.5, label="Experiment")
ax[i].plot(processed_dataset[i], linewidth=2, alpha=0.6)
ax[i].set_xlim(280, -10)
ax[i].grid()
plt.legend()
plt.tight_layout()
plt.show()
Least-squares minimization with LMFIT¶
Use the make_LMFIT_params() for a quick
setup of the fitting parameters. Note, the first two arguments of this function is
the simulator object and a list of SignalProcessor objects, processors. The
fitting parameters corresponding to the signal processor objects are generated using
SP_i_operation_j_FunctionName_FunctionArg, where i is the ith signal
processor within the list, j is the operation index of the ith processor, and
FunctionName and FunctionArg are the operation function name and function
argument, respectively.
params = sf.make_LMFIT_params(sim, processors, include={"rotor_frequency"})
print(params.pretty_print(columns=["value", "min", "max", "vary", "expr"]))
Out:
Name Value Min Max Vary Expr
SP_0_operation_1_Exponential_FWHM 20 -inf inf True None
SP_0_operation_2_Exponential_FWHM 200 -inf inf True None
SP_0_operation_4_Scale_factor 10 -inf inf True None
SP_1_operation_1_Exponential_FWHM 30 -inf inf True None
SP_1_operation_2_Exponential_FWHM 300 -inf inf True None
SP_1_operation_4_Scale_factor 100 -inf inf True None
SP_2_operation_1_Exponential_FWHM 10 -inf inf True None
SP_2_operation_2_Exponential_FWHM 150 -inf inf True None
SP_2_operation_4_Scale_factor 50 -inf inf True None
mth_0_rotor_frequency 5000 4900 5100 True None
mth_1_rotor_frequency 1940 1840 2040 True None
mth_2_rotor_frequency 960 860 1060 True None
sys_0_abundance 50 0 100 True None
sys_0_site_0_isotropic_chemical_shift 176 -inf inf True None
sys_0_site_0_shielding_symmetric_eta 0.6 0 1 True None
sys_0_site_0_shielding_symmetric_zeta 60 -inf inf True None
sys_1_abundance 50 0 100 False 100-sys_0_abundance
sys_1_site_0_isotropic_chemical_shift 43 -inf inf True None
sys_1_site_0_shielding_symmetric_eta 0.5 0 1 True None
sys_1_site_0_shielding_symmetric_zeta 30 -inf inf True None
None
Solve the minimizer using LMFIT
minner = Minimizer(sf.LMFIT_min_function, params, fcn_args=(sim, processors, sigmas))
result = minner.minimize()
result
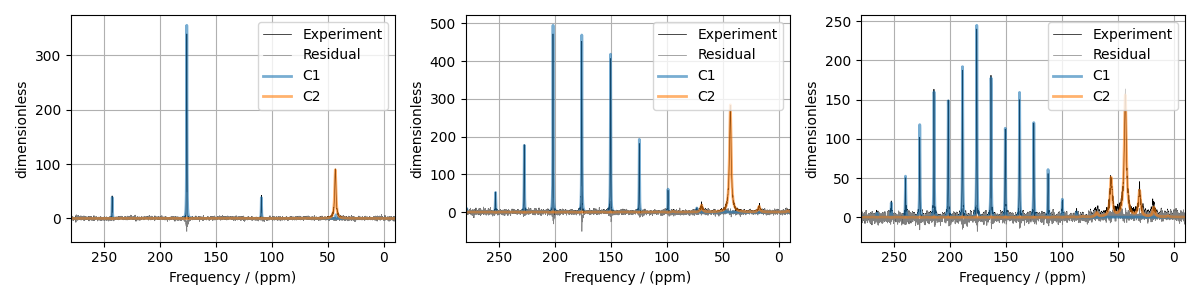
The best fit solution¶
all_best_fit = sf.bestfit(sim, processors) # a list of best fit simulations
all_residuals = sf.residuals(sim, processors) # a list of residuals
# Plot the spectrum
fig, ax = plt.subplots(1, 3, figsize=(12, 3), subplot_kw={"projection": "csdm"})
for i, proc in enumerate(processors):
ax[i].plot(experiments[i], color="black", linewidth=0.5, label="Experiment")
ax[i].plot(all_residuals[i].real, color="gray", linewidth=0.5, label="Residual")
ax[i].plot(all_best_fit[i].real, linewidth=2, alpha=0.6)
ax[i].set_xlim(280, -10)
ax[i].grid()
plt.legend()
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 11.623 seconds)
